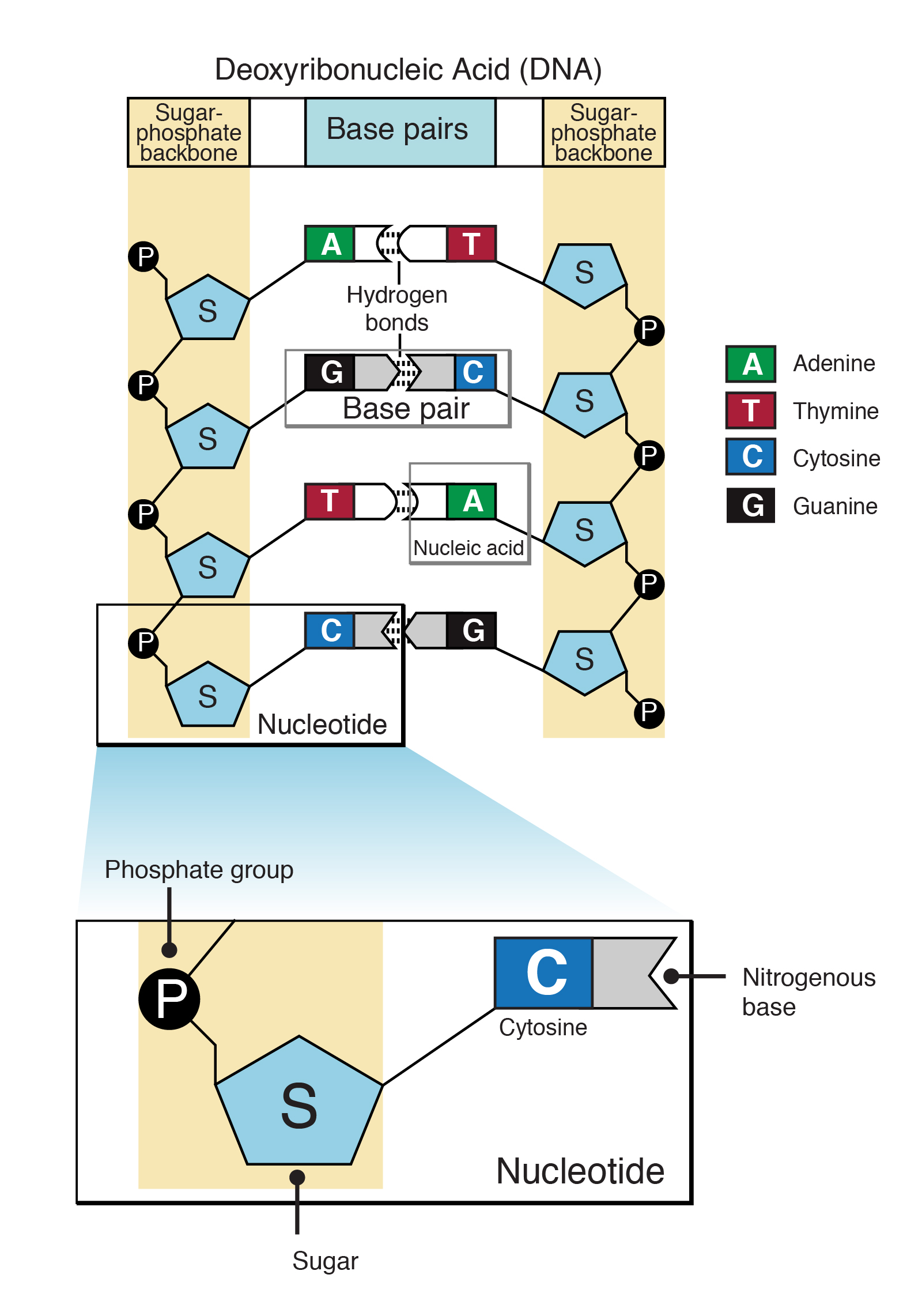
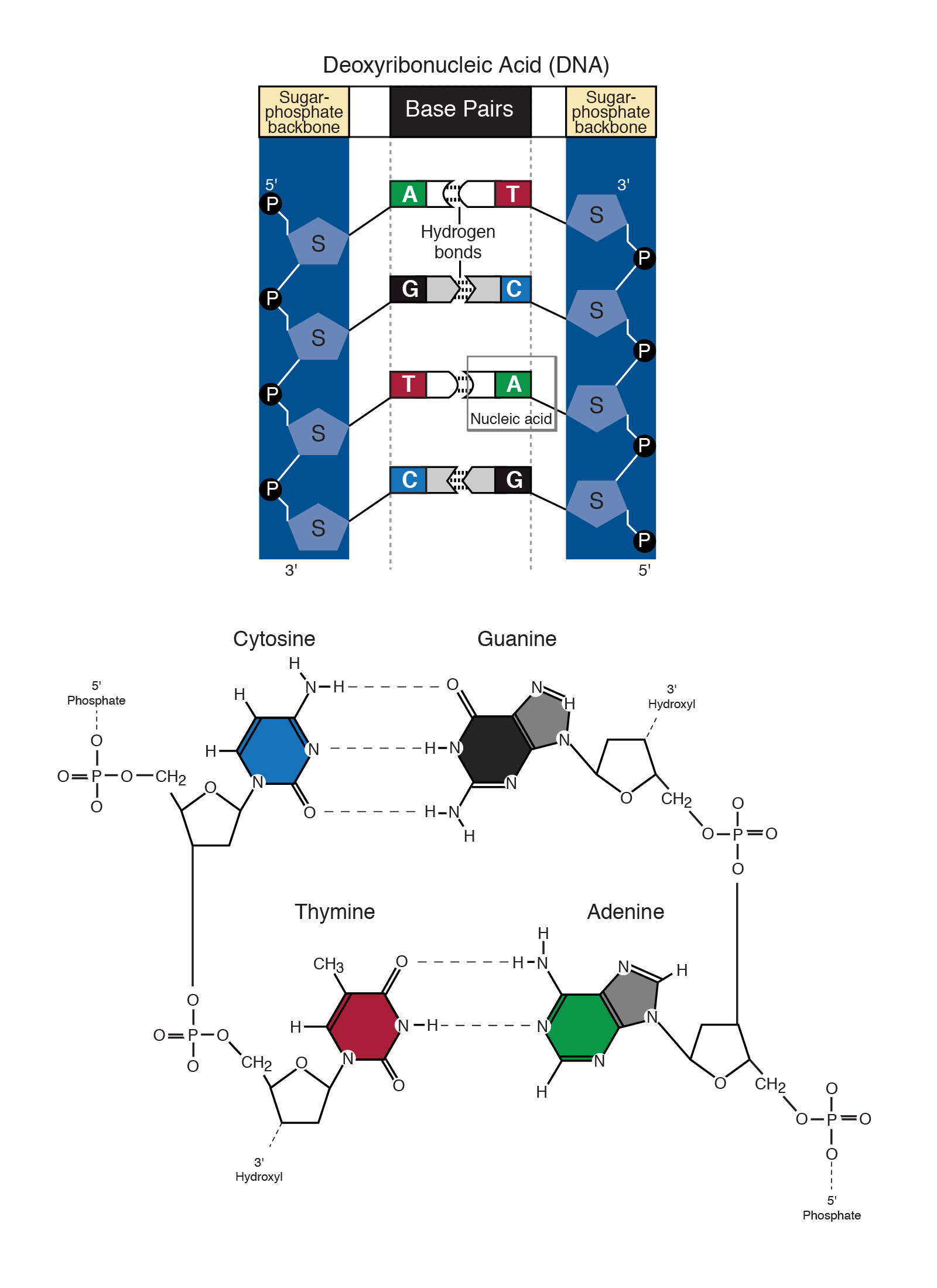
Describes How the Nucleotide Bases Are Paired Together
All As for instance must pair with Ts. B A nucleoside triphosphate is added to the 3 end.
Favorable electrostatic interactions between paired anionic and cationic amino acid sidechains are reasonably frequent in proteins.
. A A nucleoside triphosphate is added to the 5 end of the DNA releasing a molecule of pyrophosphate. Are the codons linked separately or do they overlap. 147 1 2 16 128 The read is paired mapped in a proper pair is the second read in the pair and mapped to the reverse strand.
Average or mean sequencing depth by itself eg 30 mean coverage does not take into account the percentage of bases sequenced below acceptable threshold limits or bases that were. Ion Pairs sometimes called Salt Bridges are formed when the charged group of a cationic amino acid like lysine or arginine is around 30 to 50 Å from the charged group of an anionic amino acid like aspartate. In effect total_reads 4max_id_bases reads will be compared at a time so lower numbers will produce more accurate results but consume exponentially more memory and CPU.
For the same reason you should not use -for non-interleaved paired-end data. Base pair describes the relationship between the building blocks on the strands of DNA. The other end of the nucleotide will pair up with a nucleotide holding onto the ladders other outside strand.
This is done as confirmation of preliminary clinical diagnoses evidence of carriers or. A base called adenine A links up to another called thymine T and the base guanine G pairs with cytosine C. 163 1 2 32.
The two base-pair combinations - A base-paired with T and G base-paired with C - explain the base ratios discovered by Chargaff. One end of each nucleotide holds onto an outside strand or edge of the ladder. In most cases you should probably use -at most once for an input file and at most once for an output file in order not to get mixed output.
Two primary approaches exist that allow users to interrogate RNA. There are 4 3 64 different nucleotide triplets compared with 4 2 16 possible pairs. If overlapping codons are used then fewer total nucleotides would be required.
141 1 4 8 128 The read is paired is the second read in the pair but both are unmapped. Bases nucleotide sequence has changed within the affected gene. A C T and G.
Together with other structured non-coding RNAs they perform various roles in the cell from gene regulation to translation 249. The read is paired is the second read in the pair and it is mapped while its mate is not. Cs will pair only with Gs.
Others involve insertion or deletion of one or a few nucleotides. The nucleotides are picky about who they link up with. This paper describes a new role for APE1 as the excision 35 exonuclease proofreader for DNA polymerase β particularly for removing.
The tail-n 4 prints out only the last four lines of inputfastq which are then piped into CutadaptThus Cutadapt will work only on the last read in the input file. These are the only pairs that are permissible partly because of the geometries of the nucleotide bases and the relative positions of the groups that are able to participate in hydrogen bonds and partly because the. A mutation Section 141 is a change in the nucleotide sequence of a short region of a genome Figure 141AMany mutations are point mutations that replace one nucleotide with another.
A mismatch becomes a mutation only when it is incorporated into a replicated basepaired DNA molecule. Sequentially joined triplet codons will result in a nucleotide chain three times longer than the protein it describes. Each rung contains two chemical bases bound together.
Mutations result either from errors in DNA replication or from the damaging effects of mutagens such as chemicals and radiation. So each DNA molecule is made up of two strands and there are four nucleotides present in DNA. Hydrogen bonds with the complementary nucleotide on the template strand and a phosphodiester bond with the pentose of the last incorporated nucleotide.
The minimum number of bases at the starts of reads that must be identical for reads to be grouped together for duplicate detection. A metric describing the percentage of bases sequenced across the genome or target region at a given depth eg 95 of bases covered with a minimum 10 coverage. And each of the nucleotides on one side of the strand pairs with a specific nucleotide on the other side of the strand and this makes up the.
Which of the following best describes the addition of nucleotides to a growing DNA chain.
9 1 The Structure Of Dna Concepts Of Biology 1st Canadian Edition
No comments for "Describes How the Nucleotide Bases Are Paired Together"
Post a Comment